| Gastroenterology Research, ISSN 1918-2805 print, 1918-2813 online, Open Access |
| Article copyright, the authors; Journal compilation copyright, Gastroenterol Res and Elmer Press Inc |
| Journal website https://www.gastrores.org |
Short Communication
Volume 14, Number 5, October 2021, pages 304-309
Successful Bacterial Engraftment Identified by Next-Generation Sequencing Predicts Success of Fecal Microbiota Transplant for Clostridioides difficile
Sabine Hazana, d, Sonya Daveb, Andreas J. Papoutsisa, Brad D. Barrowsa, Thomas J. Borodyc
aProgenaBiome, 1845 Knoll Drive, Ventura, CA 93003, USA
bMicrobiome Research, 1845 Knoll Drive, Ventura, CA 93003, USA
cCentre for Digestive Diseases, 229 Great North Road, Five Dock, NSW 2046, Australia
dCorresponding Author: Sabine Hazan, ProgenaBiome, 1845 Knoll Drive, Ventura, CA 93003, USA
Manuscript submitted June 14, 2021, accepted August 26, 2021, published online October 14, 2021
Short title: Assessing Success of FMT Engraftment via NGS
doi: https://doi.org/10.14740/gr1434
| Abstract | ▴Top |
Background: The effectiveness of fecal microbiota transplantation (FMT), a treatment for Clostridioides difficile infection (CDI), is dependent on successful engraftment (incorporation) of donor stool. We present a method for evaluating engraftment success based on next-generation sequencing (NGS)-based profiling of bacterial strains present in donor and recipient stool, and we suggest its potential to guide treatment decisions.
Methods: Bacterial strains in stool samples from three patients from the clinic and one donor were analyzed via NGS and metagenomic sequencing, before and 1 month after FMT for CDI. The similarity of strains present was assessed via relative abundance, principal component analysis, Shannon and Simpson diversity indexes, and Bray-Curtis dissimilarity matrix. A positive outcome was successful engraftment, where the post-FMT sample closely resembled that of the donor and CDI was cured.
Results: Patients (Pts.) 1 and 2, but not Pt. 3’s stool samples closely resembled the donor specimen post-FMT. Noteworthy, Pt. 3 pre-FMT sample was less similar to the donor than that of Pts. 1 and 2. All methods of assessing similarity and dissimilarity used yielded virtually identical conclusions. Pts. 1 and 2 which closely resembled donor specimen, eradicated CDI giving a surrogate objective measure of engraftment.
Conclusions: Success of engraftment in FMT can be assessed using NGS and metagenomic analysis and parallels success in curing CDI of the microbiome. The statistical methods we present here are reliable and consistent for such purposes. The dissimilarity of Pt. 3 to the donor combined with the failure of engraftment and failure to cure CDI in Pt. 3 suggests that FMT success may be predictable by comparing pre-FMT samples to donor. There is no clinical trial registry listing this study.
Keywords: Clostridium difficile; CDI; Donor matching
| Introduction | ▴Top |
The human intestine provides the host with a residence for tens of thousands of types of bacterial species, and this symbiotic relationship, along with the bacterial composition, is increasingly being recognized as important for human health and development [1]. It is well recognized that the gastrointestinal (GI) microbiota is not inert, but plays an active role in maturation of the gut, development of immune and mesenteric vasculature systems, and in the function of the nervous system. Imbalances in the microbiota have been reported in Clostridioides difficile infection (CDI) [1], inflammatory bowel disease [2], diabetes type II [2], obesity [2], and brain disorders [3]. Imbalances may also arise from unhealthy lifestyle or antibiotic treatment.
Fecal microbiota transplantation (FMT) is a method for altering the recipients gut microbiome, by implanting the microbiome of a healthy donor into the gut of an unhealthy recipient. FMT is accepted as standard therapy for toxigenic CDI, one of its most common indications [1]. FMT success depends mainly on successful engraftment [4], the process of acceptance and growth of transplanted fecal bacteria.
Routes of transplantation for FMT include oral capsules, enema or colonoscopy for lower GI tract, and upper endoscopy or nasogastric tube infusion for upper GI tract [1]. Although fecal enemas are relatively easy and inexpensive, they can result in limited distribution of fecal matter. Colonoscopy, on the other hand, can be effective at distributing stool throughout the colon, and possibly terminal ileum [1]. Additionally, colonoscopy may be less likely to require multiple attempts for successful FMT. Thus, colonoscopy is generally the preferred method of FMT, though there have not been controlled comparative studies [1].
While FMT is a standard method for CDI treatment, factors governing success remain unclear; and there is still at least an 8% failure rate for CDI, and much higher failure rates for other diseases [4]. Host-donor matching is known to play a role in engraftment success for CDI and other diseases [5, 6]. As reviewed by Wilson et al [4], success depends on the composition, and especially diversity, of the microbiome in the donor, and as such certain donors could have particularly engraftable feces. In short, careful selection of a donor may impact engraftment success and thus clinical efficacy.
The ability to measure engraftment can be useful for guidance in donor selection and in evaluating success of FMT therapy. Engraftment could be measured as similarity of species between stools of recipient and donor, post-FMT. Next-generation sequencing (NGS) represents an efficient high-throughput method of sequencing deoxyribonucleic acid (DNA) [7], including bacterial DNA in stool. Metagenomic shotgun analysis [8] allows this sequence information to be analyzed for determining strains of bacteria present. We present a method by which NGS was successfully used to sequence donor and recipient stool microbial DNA, with bacterial species level resolution, and thereby distinguish successful from unsuccessful transplant engraftments. We also observed lack of engraftment and clinical success only in the patient most divergent in pre-FMT diversity from the donor.
| Materials and Methods | ▴Top |
Fecal transplant methods
FMT transplant material (TM) was obtained from healthy donor stool bank and stored in -80 °C freezer until the transplant procedure. All three patients were scheduled for FMT based on recurrence of CDI. After third recurrence, patients were given vancomycin 500 mg orally three times a day for 10 days. On day 11, prior to colonoscopy, they were given a standard bowel preparation. Colonoscopy was performed with patients receiving propofol. The colonoscope was inserted to the cecum, and 300 mL of TM was infused directly into the cecum avoiding the terminal ileum. Patients recovered in Trendelenburg position for 30 min to pool the TM in the proximal colon, followed by 2 mg of Lomotil and 10 mg of Bentyl to hold solution longer. Post-FMT, patients were told to avoid greens and dairy for 1 month.
NGS
First, NGS was performed (n = 7) on fecal samples from three study participants and one donor sample from one donor individual, using the methods outlined previously [9]. Patient stools were collected both prior to and 1 month after FMT. From the stool specimens, DNA was extracted utilizing the Qiagen PowerFecal DNA Kit from 200 mL of stool material that was suspended in the DNA/ribonucleic acid (RNA) Shield stabilization solution present in the Zymo collection vials. Zymo’s DNA/RNA Shield is designed to preserve nucleic acid genetic integrity and prevent degradation at ambient temperature for > 1 month, or > -20 °C indefinitely purified DNA was then quantified and normalized for downstream library fabrication utilizing shotgun methodology [8]. Prepared and indexed libraries were subsequently pooled and sequenced with Illumina’s NextSeq 500/550 High-Output v2.5 300 cycle kiton the Illumina NextSeq 550 System.
Next, DNA sequences were analyzed and compared, in terms of species present, in recipients vs. donors using metagenomic sequencing analysis. Sample FASTQ files were analyzed with downstream bioinformatics to profile the microbial communities from metagenomic sequencing data. Specifically, sequencing acceptance criteria were a Q-score (AQ30) 75%, cluster density between 120 and 240 K/mm2, and clusters passing filter (PF%) 80%. Following successful NGS QC, sequences were then mapped utilizing the sequencing alignment tool in One Codex’s bioinformatics analysis pipeline. Diversity analyses were performed to characterize the intestinal microbiota of the study participants. The metagenomic analyses included the principal component analysis (PCA), Bray-Curtis dissimilarity matrix, and the Simpson and Shannon diversity indexes.
Ethics statements
All subjects provided informed consents. This study was approved by New England Institutional Review Board. This study was conducted in compliance with common ethical standards on human subjects and with the Helsinki Declaration.
| Results | ▴Top |
Seven samples were sequenced (the donor and three patients, each before and after transplant), then species present were compared between donor and recipient. Higher similarity of recipient species population post-FMT to the donor is indicative of a more successful engraftment.
Figure 1 shows relative abundance of bacteria in samples, organized by phylum and family. Phylum Firmicutes was the most abundant, with a relative abundance of 73% (donor), and in pre-FMT samples, 51% (Patient 1 (Pt. 1)), 15% (Pt. 2), and 43% (Pt. 3) of bacteria. After transplant, these relative abundances more closely resembled the donor at 53% for Pt. 1 and 60% for Pt. 2, but less closely at 16% for Pt. 3. Family Lachnospiraceae was present at 15% for donor, and pre-FMT, for Pts. 1, 2, and 3 at 34%, 1.7%, and 23%, respectively. Post-FMT Lachnospiraceae was present, in Pts. 1, 2 and 3, at 19%, 27%, and 0.36%, respectively. Thus, for the two most abundant families, after FMT, Pts. 1 and 2 more closely resembled the donor, while Pt. 3 did not, suggesting Pts. 1 and 2, but not Pt. 3, had successful engraftments.
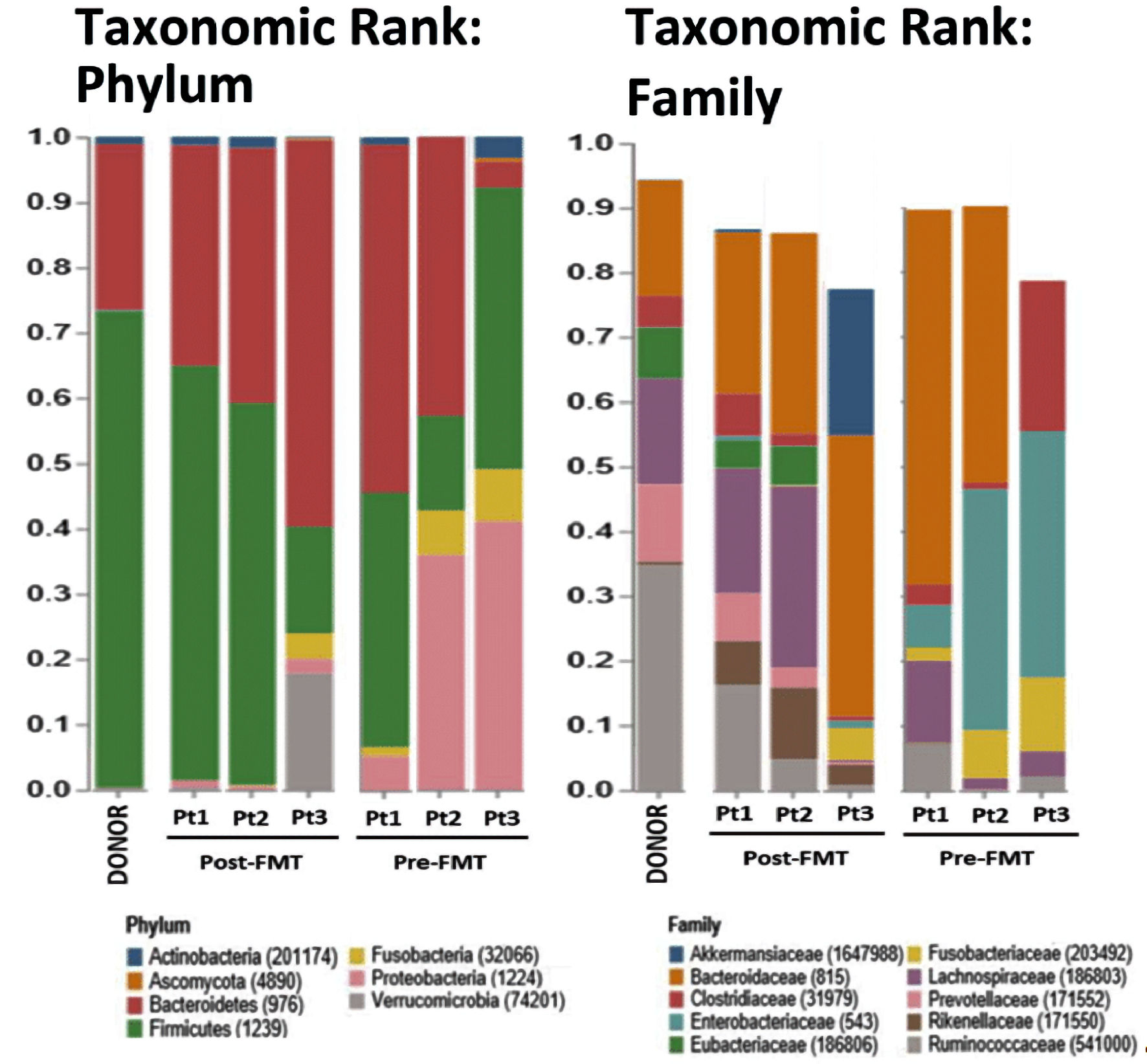 Click for large image | Figure 1. Relative abundances (RAs, %) of bacteria in stool samples of donor, post-FMT and pre-FMT samples. Columns represent individual patients with colored bars capturing their respective RAs (%) at the phylum and family levels. The RAs of phylum Firmicutes in the donor, patient (Pt.) 1, Pt. 2, and Pt. 3 post-FMT were 73%, 63%, 60%, and 16%, respectively; as compared to the patients matched pre-FMT of 51%, 15%, and 43%. The relative abundance of family Lachnospiraceae in the donor, Pt. 1, Pt. 2, and Pt. 3 post-FMT were 15%, 19%, 27%, and 0.36%, respectively; as compared to the patients matched pre-FMT RAs of 34%, 1.7%, and 23%, respectively. |
Next, PCA was used to compare species present in the seven samples. PCA is a method of grouping data based on similarity. In this case, it was similarity of total species present in the fecal samples. Data that appear closer together on PCA graphs are more similar, while data further apart are less similar. Figure 2 shows such a PCA graph, with a square for donor, circles for Pre-FMT patients, and color-matched triangles for post-FMT patient samples. As highlighted, post-FMT, Pts. 1 and 2 are relatively similar to donor (closer on PCA graph), while Pt. 3 is less similar. Thus, PCA also supports that Pts. 1 and 2 are successful engraftments.
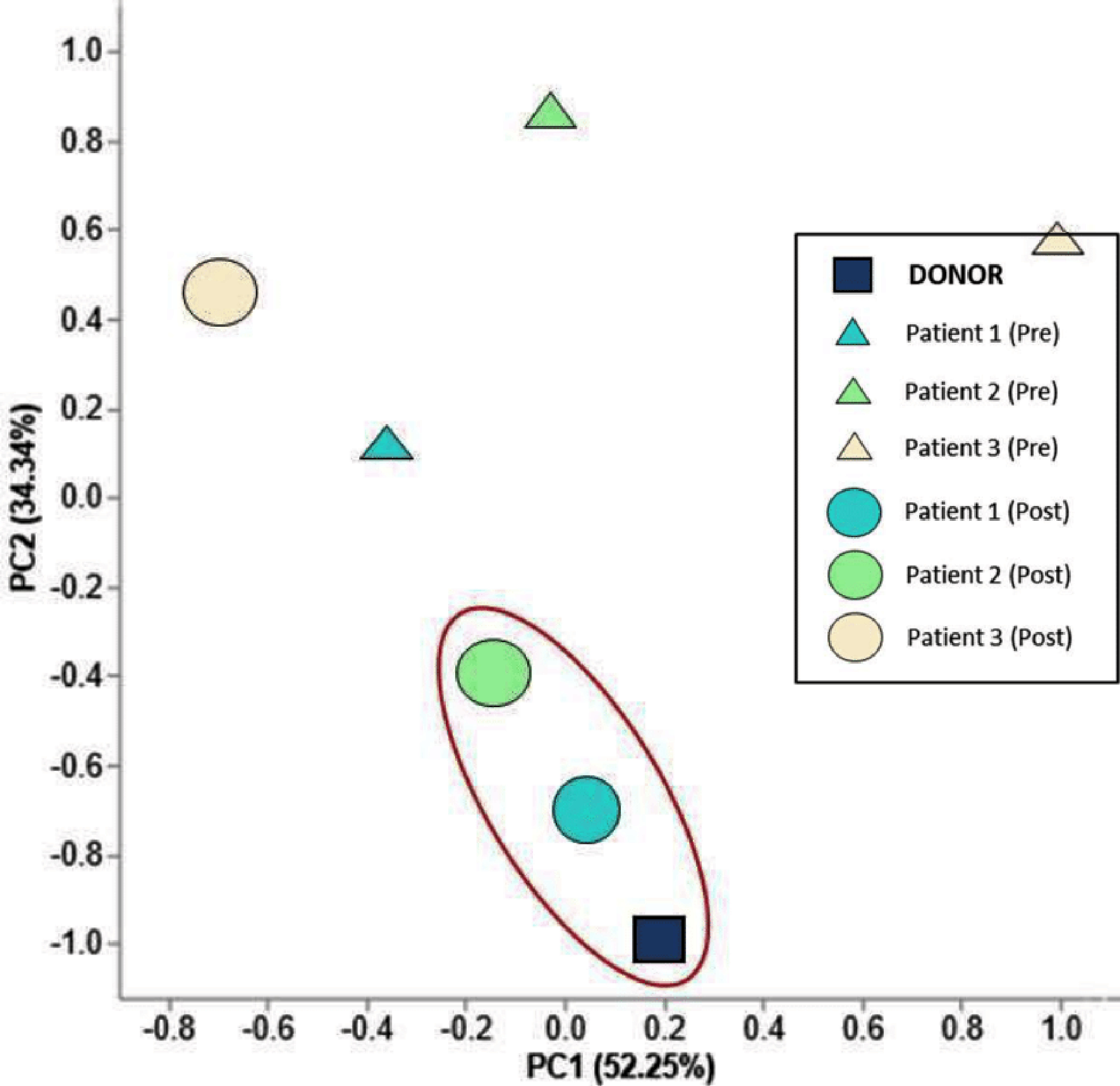 Click for large image | Figure 2. Principal component analysis (PCA) of the microbiota from the donor, pre-FMT patients, and post-FMT patients. Axes are labeled with the percent of variance they explain. Triangles represent pre-FMT, circles represent post-FMT, and the square represents FMT donor. PC1 captures 52.25% of the variation and elucidates the shift of the two successful FMTs compared to their microbiota prior to engraftment. Samples circled in red highlight the patient similarity following successful FMTs with that of donor stool. |
Diversity indices, which represent the amount and diversity of species present in a sample, were also used to compare the patients with the donor before and after FMT. Figure 3 shows two common indexes utilized in metagenomic analyses, the Shannon and Simpson [10], for the samples. The Shannon index of the donor, and Pt. 1, Pt. 2, and Pt. 3 following FMT were 6.0, 6.0, 5.5, and 3.4, respectively. The Simpson index of the donor, and Pt. 1, Pt. 2, and Pt. 3 post-FMT were 0.89, 0.89, 0.84, and 0.72, respectively. Collectively, this data illustrate that for Pt. 1 (green) and Pt. 2 (blue), the diversity indices were more similar to the donor (black) post-FMT than pre-FMT, while for Pt. 3, the species diversity was less similar to the donor post-FMT. Thus, these diversity indices also support that Pts. 1 and 2, but not Pt. 3, are successful engraftments.
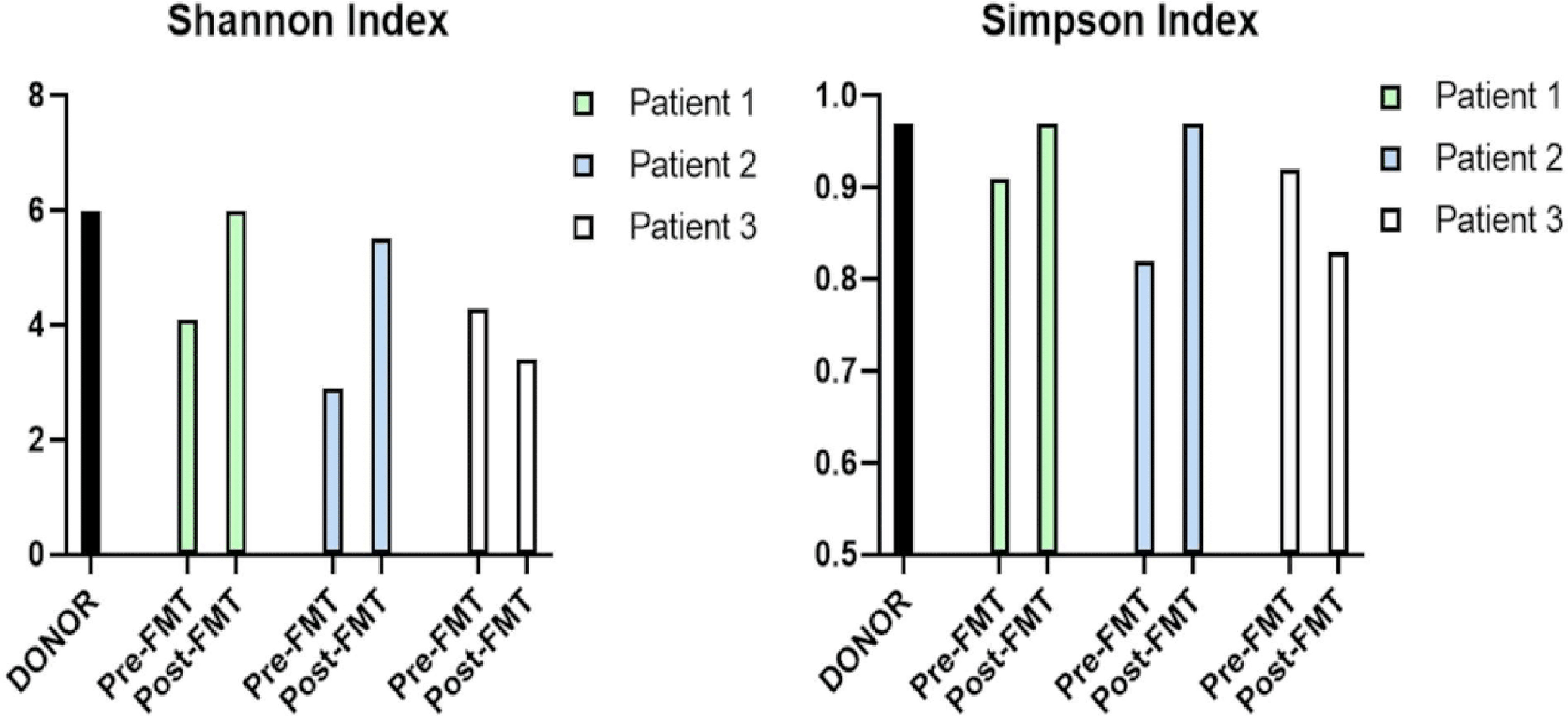 Click for large image | Figure 3. Alpha diversity of the microbiota from the donor, pre-FMT and post-FMT samples for the Shannon and Simpson indexes. Both indexes for patients (Pts.) 1 and 2 match their donor’s, whereas Pt. 3 does not, indicative of an unsuccessful engraftment. The Shannon index of the donor, Pt. 1, Pt. 2, and Pt. 3 following FMT were 6.0, 6.0, 5.5, and 3.4, respectively. The patients matched pre-FMTs Shannon indexes were 4.1, 2.9, and 4.3, respectively. The Simpson index of the donor, Pt. 1, Pt. 2, and Pt. 3 post-FMT were 0.89, 0.89, 0.84, and 0.72, respectively. The patients matched pre-FMTs Simpson indexes were 0.91, 0.82, and 0.92. FMT: fecal microbiota transplantation. |
Figure 4 depicts the Bray-Curtis distance matrix for the seven samples, with dark blue indicative of similarity and lighter blue of dissimilarity. Looking at the row labeled “Donor”, the corresponding column of “Pt. 1 (post)” is dark blue, consistent with high similarity and successful engraftment. Conversely, the column of “Pt. 3 (post)” is light blue, indicative of less similarity and consistent with poor engraftment. This analysis demonstrates Pt. 1 as the most, and Pt. 3 least, engraftment success.
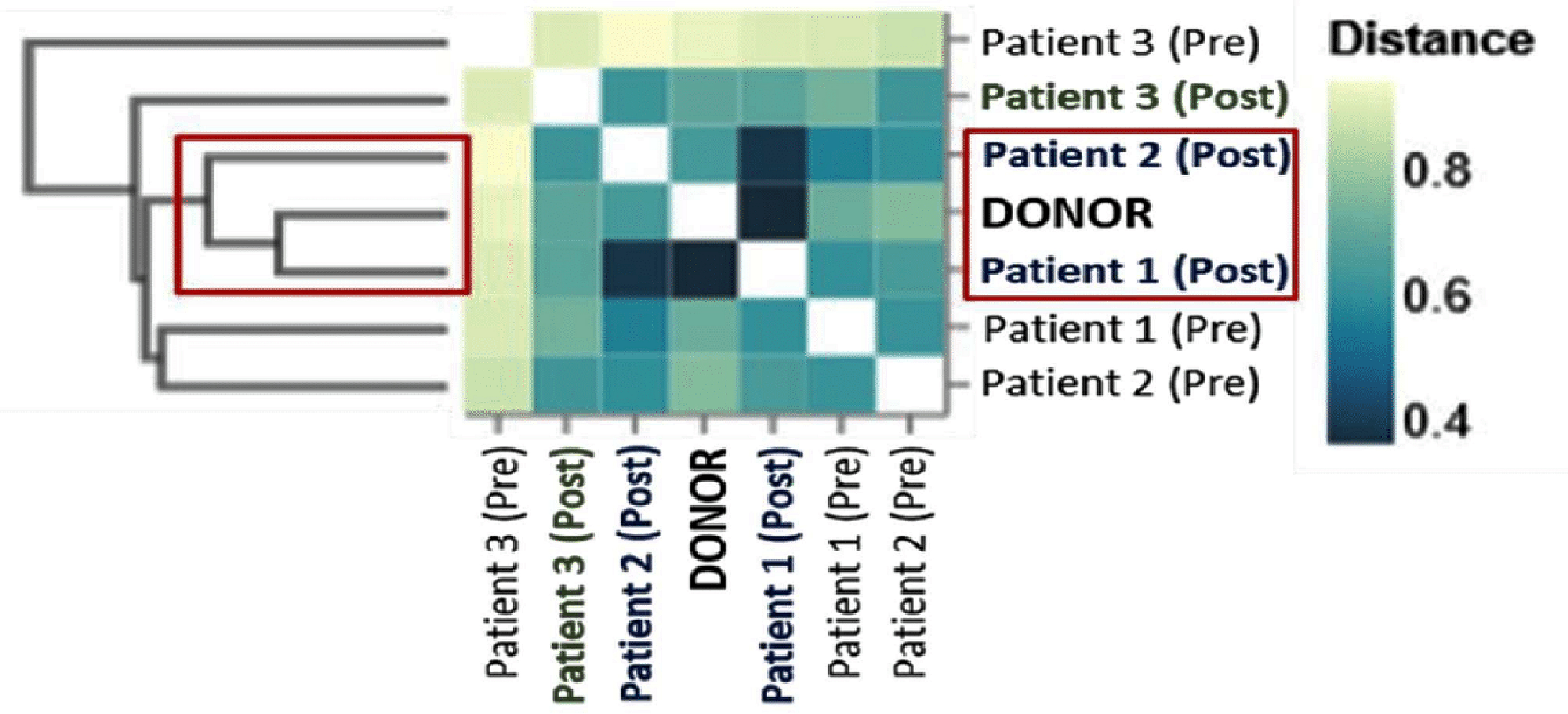 Click for large image | Figure 4. Bray-Curtis distance matrix of the microbiota from donor, pre-FMT and post-FMT samples. The ordination plots are based on the bacterial profiles for each. The Bray-Curtis matrix identifies the similarity of patients (Pts.) 1 and 2 after fecal microbiota transplantation (FMT) with that of the donor (dark blue squares), clustering in distinct coordinates of the matrix. It also identifies that Pt. 3’s ordination is the most dissimilar of the three post-FMT samples. This provides insight that Pt. 3 may have had an unsuccessful FMT, whereas Pts. 1 and 2 are more likely to have had successful engraftments. |
| Discussion | ▴Top |
We present preliminary results of a method for quantifying success of FMT engraftment using NGS and metagenomic analysis, which we applied at a time period of 1 month. NGS is considered the best available quantitative approach for profiling microbiomes [7]. Given the ease and speed of NGS (a bacterial genome can be sequenced in 1 day [7]), as well as the automation possible for such standard statistical analysis, this method can be extremely valuable as a rapid tool for quantifying FMT success and profiling donor and recipients in order to guide treatment decisions. Of course the initial costs of new technologies are high when utilized at a low rate (our machine reads 20 samples simultaneously). However, as technologies of any kind become more popular, their economic practicality also increases.
We present a thorough confirmation of engraftment success by five different established analytical methods: 1) relative abundance; 2) PCA; 3) Shannon’s index; 4) Simpson’s index and 5) Bray-Curtis matrix. Regardless of the method utilized, we obtain the same result; Pts. 1 and 2 engrafted successfully, and Pt. 3 did not. Thus, NGS combined with metagenomic shotgun analysis is a reliable method for comparing species present in fecal samples to support successful engraftment. In light of the consistency seen, these may be some of the most appropriate analytical methods for predicting engraftment success.
Why did Pt. 3 not graft successfully, when given the same donor sample as the other two patients? The pre-FMT data (especially Figs. 1, 2, and 4) showed Pt. 3 bacterial strain composition was the most dissimilar from donor. Donor-recipient matching is often considered to be helpful for FMT success. However, until now there was no technology to tell us that Pt.3 would not be successfully cured of CDI by FMT using the present donor. If one were to profile, by methods we suggest here, the bacteria in the pre-FMT samples and donors, one could search for matching donors to increase likelihood of FMT success.
This study was limited by the small sample size, and information from more subjects would be informative. With a larger sample size, one could come up with a cut-off for the similarity measures used (e.g., Shannon and Simpson indices, Bray-Curtis distance matrix) predicting whether a certain level of similarity will or will not lead to clinical success.
In conclusion, the treatment of CDI and possibly other diseases via FMT can be made more effective by profiling the strains present prior to performing the transplant. Ideally, one can screen patients and donors prior to the FMT procedure and select the best donor sample. Additionally, one can profile the post-FMT stool samples, to evaluate treatment success, and alter future treatment methods if needed. For a rapidly spreading and life-threatening disease like CDI, this analysis of compatibility can be critical for timely patient treatment.
Acknowledgments
None to declare.
Financial Disclosure
This study was funded by ProgenaBiome, LLC and Dr. S. Hazan.
Conflict of Interest
Drs. S. Hazan, B. Barrows, A.J. Papoutsis and T. Borody have corporate affiliation to ProgenaBiome™. Dr. S. Hazan has additional corporate affiliation to Ventura Clinical Trials and Topelia Therapeutics Inc. Dr. T. Borody has additional corporate affiliation to Topelia Therapeutics Inc. Dr. S. Dave has corporate affiliation to North End Advisory, LLC and McKesson Specialty Health. No products of these affiliated companies were used in this study.
Informed Consent
All subjects provided informed consents.
Author Contributions
Dr. S. Hazan led and contributed to all aspects of the study and writing. Dr. S. Dave wrote the manuscript and edited it. Dr. T.J. Borody edited and Drs. A.J. Papoutsis and B.D. Barrows carried out the metagenomic analyses. Dr. A.J. Papoutsis performed the sequencing, wrote the initial poster and outline of study.
Data Availability
The data supporting the findings of this study are available from the corresponding author upon reasonable request.
Abbreviations
CDI: Clostridioides difficile infection; FMT: fecal microbiota transplantation; NGS: next-generation sequencing; Pt.: patient; PCA: principle component analysis; RAs: relative abundances; TM: transplant material
| References | ▴Top |
- Jung Lee W, Lattimer LD, Stephen S, Borum ML, Doman DB. Fecal microbiota transplantation: a review of emerging indications beyond relapsing clostridium difficile toxin colitis. Gastroenterol Hepatol (N Y). 2015;11(1):24-32.
- Bull MJ, Plummer NT. Part 1: The human gut microbiome in health and disease. Integr Med (Encinitas). 2014;13(6):17-22.
- Zhu S, Jiang Y, Xu K, Cui M, Ye W, Zhao G, Jin L, et al. The progress of gut microbiome research related to brain disorders. J Neuroinflammation. 2020;17(1):25.
doi pubmed - Wilson BC, Vatanen T, Cutfield WS, O'Sullivan JM. The super-donor phenomenon in fecal microbiota transplantation. Front Cell Infect Microbiol. 2019;9:2.
doi pubmed - Ng SC, Kamm MA, Yeoh YK, Chan PKS, Zuo T, Tang W, Sood A, et al. Scientific frontiers in faecal microbiota transplantation: joint document of Asia-Pacific Association of Gastroenterology (APAGE) and Asia-Pacific Society for Digestive Endoscopy (APSDE). Gut. 2020;69(1):83-91.
doi pubmed - Duvallet C, Gibbons SM, Gurry T, Irizarry RA, Alm EJ. Meta-analysis of gut microbiome studies identifies disease-specific and shared responses. Nat Commun. 2017;8(1):1784.
doi pubmed - Goldberg B, Sichtig H, Geyer C, Ledeboer N, Weinstock GM. Making the leap from research laboratory to clinic: challenges and opportunities for next-generation sequencing in infectious disease diagnostics. mBio. 2015;6(6):e01888-01815.
doi - Sharpton TJ. An introduction to the analysis of shotgun metagenomic data. Front Plant Sci. 2014;5:209.
doi pubmed - Papoutsis A, Borody T, Dolai S, Daniels J, Steinberg S, Barrows B, Hazan S. Detection of SARS-CoV-2 from patient fecal samples by whole genome sequencing. Gut Pathog. 2021;13(1):7.
doi pubmed - Wagner BD, Grunwald GK, Zerbe GO, Mikulich-Gilbertson SK, Robertson CE, Zemanick ET, Harris JK. On the use of diversity measures in longitudinal sequencing studies of microbial communities. Front Microbiol. 2018;9:1037.
doi pubmed
This article is distributed under the terms of the Creative Commons Attribution Non-Commercial 4.0 International License, which permits unrestricted non-commercial use, distribution, and reproduction in any medium, provided the original work is properly cited.
Gastroenterology Research is published by Elmer Press Inc.


