| Gastroenterology Research, ISSN 1918-2805 print, 1918-2813 online, Open Access |
| Article copyright, the authors; Journal compilation copyright, Gastroenterol Res and Elmer Press Inc |
| Journal website http://www.gastrores.org |
Original Article
Volume 10, Number 2, April 2017, pages 100-105
Assessment of the Relation Between SNP in MxA Gene and the Responsiveness of Egyptian HCV Genotype 4 Patients to Pegylated Interferon and Ribavirin Treatment
Mohamed Hassanya, c, Amany Gamalb, Nashwa Zakib, Basem Eysaa
aTropical Medicine Department, National Hepatology and Tropical Medicine Research Institute, Cairo, Egypt
bDepartment of Clinical Pathology, National Hepatology and Tropical Medicine Research Institute, Cairo, Egypt
cCorresponding Author: Mohamed Hassany, Tropical Medicine Department, National Hepatology and Tropical Medicine Research Institute, Cairo, Egypt
Manuscript accepted for publication March 10, 2017
Short title: SNP in MxA Gene and Responsiveness to PegIFN/RBV
doi: https://doi.org/10.14740/gr810w
| Abstract | ▴Top |
Background: Pegylated interferon (PegIFN) is used in the treatment of chronic hepatitis C virus (HCV) patients especially in resource limited countries. Treatment with PegIFN stimulates the expression of a number of host genes encoding enzymes with antiviral activities, including myxovirus resistance gene-A (MxA gene). MxA gene was found to have a single nucleotide polymorphism (SNP) at position -88 in the promoter region that affects the expression of MxA gene protein and was suggested to affect the treatment outcome. The aim of the work was to assess the relation between the SNP in the MxA gene and its impact on treatment of chronic HCV patients with PegIFN and ribavirin.
Methods: We therefore genotyped the biallelic G/T SNP in the promoter region of MxA gene at position -88 from the transcription start site by restriction fragment length polymorphism (RFLP) in 70 chronic HCV genotype 4 interferon naive Egyptians and 40 healthy controls.
Results: G allele was the prevalent one in both HCV patients group (105, 74.5%) and control group (66, 82.5%), while T allele was less expressed in patients group (36, 25.5%) and control group (14, 17.5%). There is no correlation between genotypes and response to IFN-alpha therapy: GG (OR: 0.958, 95% CI: 0.541 - 1.698, P = 0.884), GT (OR: 0.667, 95% CI: 0.188 - 2.362, P = 0.530), and TT (OR: 0.300, 95% CI: 0.083 - 1.090, P = 0.067).
Conclusion: MxA nt-88 SNP did not affect the sustained virological response (SVR) rates after PegIFN and ribavirin combined treatment and did not act as a biological marker to potentially identify responders and non-responders to treatment. Our results call for additional large studies and/or meta-analysis of all currently available data to examine the role of MxA nt-88 SNP in predicting response to PegIFN and ribavirin in patients with IFN-alpha naive HCV genotype 4.
Keywords: HCV; MxA gene; Pegylated interferon
| Introduction | ▴Top |
Hepatitis C virus (HCV) related chronic liver disease remains one of the most important global health problems. Egypt still has a very high prevalence of chronic HCV even after running for many years in building a national treatment program; prevention and screening plan leads to decline of HCV infection from 14.7% (antibodies prevalence) and 10% (viremic patients) in age group of 15 - 59 years in demographic and health survey (DHS) in 2008 [1] to 10% (antibodies prevalence) and 7% (viremic patients) in DHS in 2015 in the same age group [2].
Evolution of HCV treatment with direct acting antivirals (DAAs) leads to improvement of treatment outcome in all genotypes [3], yet the very high costs of DAAs drugs make it unavailable in many countries especially in resource limited countries, rendering the treatment with pegylated interferon (PegIFN) and ribavirin (RBV), the corner stone of treatment in these countries due to its relative low costs; the European Association for the Study of the Liver (EASL) considered the treatment with PegIFN + RBV as an alternative option in case of non-availability of DAAs [4].
Predictors of treatment of response to PegIFN include several host and viral related factors: searching for genetic polymorphism which may lead to improvement of treatment outcome with PegIFN is the overriding concern for many scientists; IL28B gene mutation was found to be a good predictor for achieving sustained viral response (SVR) as noticed in many trials, showing favorable outcome with IL28B CC genotype higher than those carrying CT or TT genotypes [5]. The myxovirus resistance gene-A (MxA) is a protein that is induced by IFN and influences the IFN-induced antiviral activities of host cells against viruses [6]; it was found to have a single nucleotide polymorphism (SNP) at position -88 in the promoter region that affects the expression of MxA protein and could affect HCV response to treatment. The levels of the MxA protein were greater in virological responders than in non-responders (NR) [7]. The SNPs in the promoter region of the MxA gene were found to be most likely associated with the levels of IFN-induced expression of the MxA protein, and thus further with the response of the hepatitis C patients to the IFN therapy [8].
Aim of the study
The aim of the study was to evaluate the possible relation between MxA promoter -88 SNP and the response to treatment with PegIFN and RBV in chronic HCV genotype 4 patients.
| Patients and Methods | ▴Top |
The study was reviewed and approved by Independent Ethics Committees and conducted in accordance with the Declaration of Helsinki and Good Clinical Practice guidelines. All enrolled patients provided written, informed consent prior to the start of the study.
Seventy chronic HCV genotype 4 interferon naive patients were enrolled through the national treatment program at hepatology clinic, National Hepatology and Tropical Medicine Research Institute (NHTMRI), Cairo between the period (2014 - 2015) and end of follow-up period in early 2016. The patients received treatment with weekly PegIFN alpha 2a and daily weight based RBV (1,000 mg/day for those < 70 kg, and 1,200 mg/day for those ≥ 70 kg) and then were classified into two groups: group 1 (sustained-responders group, 40 patients) received 48 weeks of treatment and achieved SVR (defined as HCV RNA negativity by polymerase chain reaction testing (PCR) after 24 weeks of treatment stoppage) and group 2 (non-responders group, 30 patients) received 12 weeks of treatment and failed to get any decline in viral load.
A control group composed of 40 persons of equally distributed sex (20 males and 20 females) was also included in the study.
Screening for polymorphisms
The host DNA was used for amplification of the Mx1 gene by traditional PCR. The biallelic G/T polymorphism in the promoter region of Mx1 at position -88 from the transcription start site was genotyped by RFLP using the enzyme HhaI (New England Biolabs) to digest the PCR fragment of 351 bp. Amplification was carried out in a volume of 20 μL, containing 100 ng DNA, 2.5 mM MgCl2 (Promega, USA), 500 nM of each primer (Eurofins, UK); primers were: #MXAF01 (5-ACACACCCGTTTCCACCCTGGAGAGGCCAG-3), forward, and #MXAR02 (5-TGCGCAGTGCTGGAGTGCGGCCTCCGCTCT-3), reverse, 500 mM dNTP’s (Promega, USA), 1x PCR buffer (Promega, USA), and 1 U GoTaq DNA polymerase enzyme (Promega, USA). The cycling conditions in an applied biosystems 9700 machine were: denaturation at 94 °C for 5 min, subsequently 35 cycles of denaturation at 94 °C for 30 s; annealing at 58 °C for 30 s; and extension at 72 °C for 1 min. This was followed by a final extension step at 72 °C for 7 min. For the HhaI restriction digest 8 μL of the PCR product was digested for at least 4 h in a volume of 20 μL with 5 U of HhaI according to the manufacturer’s specifications. Digested PCR product (10 μL) was run out on 2% agarose gels and analyzed. In the presence of the G allele, the 351 bp long product was cut into fragments of 261, 51, 23 and 16 bp and in the presence of the T allele into fragments of 312, 23 and 16 bp.
Statistical analysis
Analysis of data was performed using SPSS 21 for Windows. Description of variables was presented as follows. Description of numerical variables was in the form of mean, and standard deviation (SD), and description of categorical variables was in the form of numbers (No.) and percent (%).
Numerical data were not normally distributed. Accordingly, non-parametric tests were used for comparison. This was carried out by Mann-Whitney U test when comparing between two groups of independent variables. Kruskal-Wallis test was used when comparing between more than two groups of independent variables. Results were expressed in the form of P-values.
Comparison between categorical variables was carried out by Chi-square test (χ2).
The significance of the results was assessed in the form of P-value that was differentiated into: 1) P-value of > 0.05 which is considered statistically not-significantly different, and 2) P-value of ≤ 0.05, which is considered statistically significantly different.
Standard univariable analysis was conducted using contingency tables. Odds ratios (ORs) and 95% confidence intervals (95% CIs) were calculated along with maximum likelihood and Fisher’s exact P-values, as appropriate.
| Results | ▴Top |
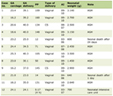
Tables 1 and 2 show the baseline demographic, clinical and laboratory data of the studied groups; the mean age of the studied patients was 45.23 ± 7.10 years, 50 patients (71.5%) were males, while 20 patients (28.5%) were females; 52 patients (74.2%) had a fibrosis score (F1) according to Metavir score [9], 12 patients (17.2%) had F2, while six patients (8.6%) had F3. No significant difference was observed between non-responders and sustained responders regarding age, ALT, AFP levels or HCV viral load. The dominant allele of MxA gene found in this study was GG polymorphism, which was expressed in 47 patients (67%), followed by TT in 13 patients (18.5%), and then lastly GT (14.5%); in the control group, the dominant allele was also GG in 30 persons (75%), then GT in six persons (15%), and then TT in four persons (10%) as shown in Table 3.
 Click to view | Table 1. Baseline Demographic and Laboratory Data of Studied Patients |
 Click to view | Table 2. Demographic and Laboratory Data in Non-Responders and Sustained Responders Groups |
 Click to view | Table 3. Distributions of the MxA Promoter Genotypes Among Studied Patients |
Table 4 and Figure 1 show that there is no correlation between genotypes and response to IFN-alpha therapy: GG (OR: 0.958, 95% CI: 0.541 - 1.698, P = 0.884), GT (OR: 0.667, 95% CI: 0.188 - 2.362, P = 0.530), and TT (OR: 0.300, 95% CI: 0.083 - 1.090, P = 0.067).
 Click to view | Table 4. Genotyping and Allele Frequency of Patients and Control Groups |
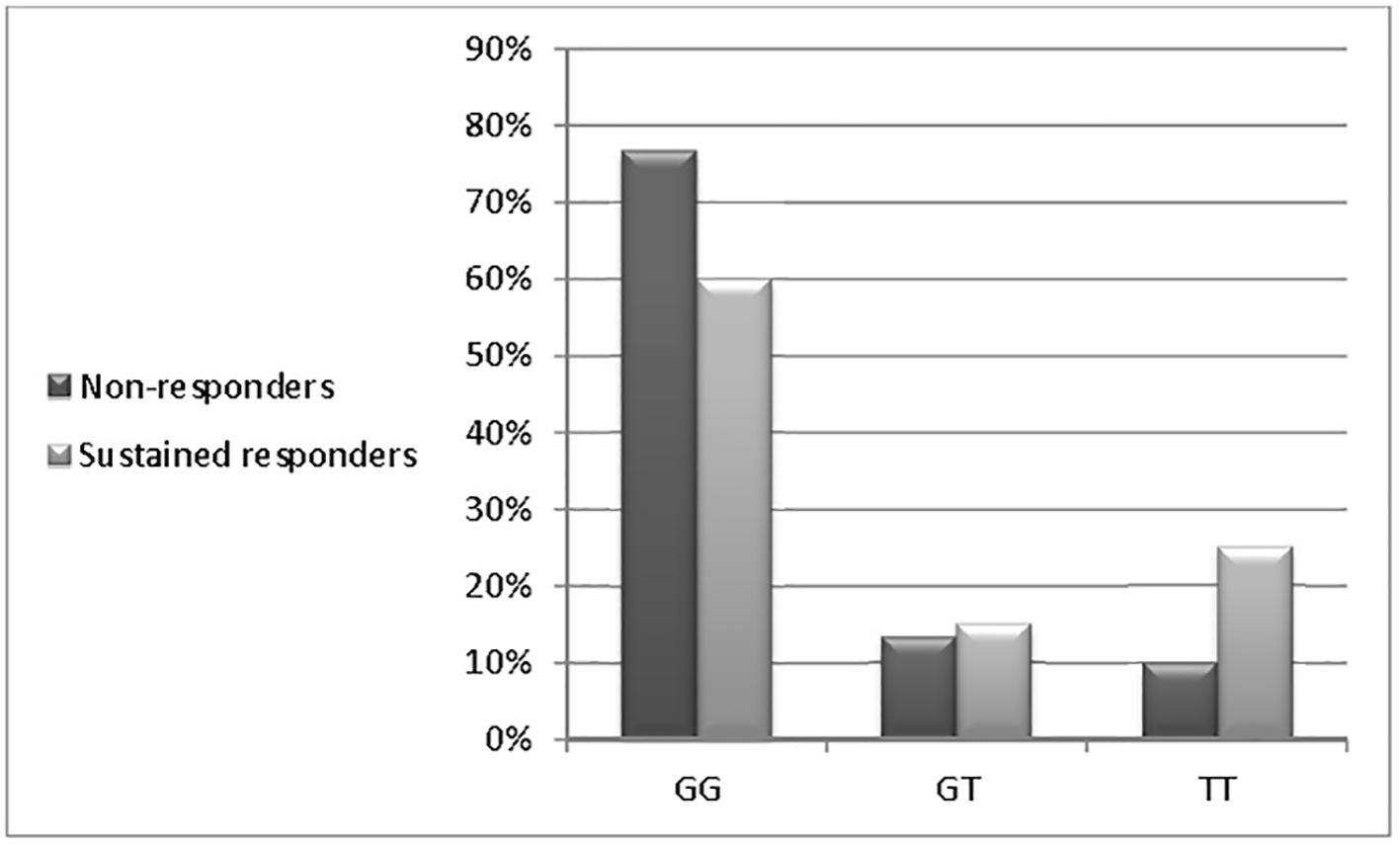 Click for large image | Figure 1. Distributions of the MxA promoter genotypes among studied patients. |
Table 3 and Figure 2 show that G allele is the prevalent one in both HCV patients group (105, 74.5%) and control group (66, 82.5%), while T allele was less expressed in patients group (36, 25.5%) and control group (14, 17.5%). In other tested parameters, the response to treatment was not affected by age, gender or ALT levels, meanwhile all patients with advanced fibrosis and necroinflammation (A3/F3) were non-responders as shown in Tables 5 and 6.
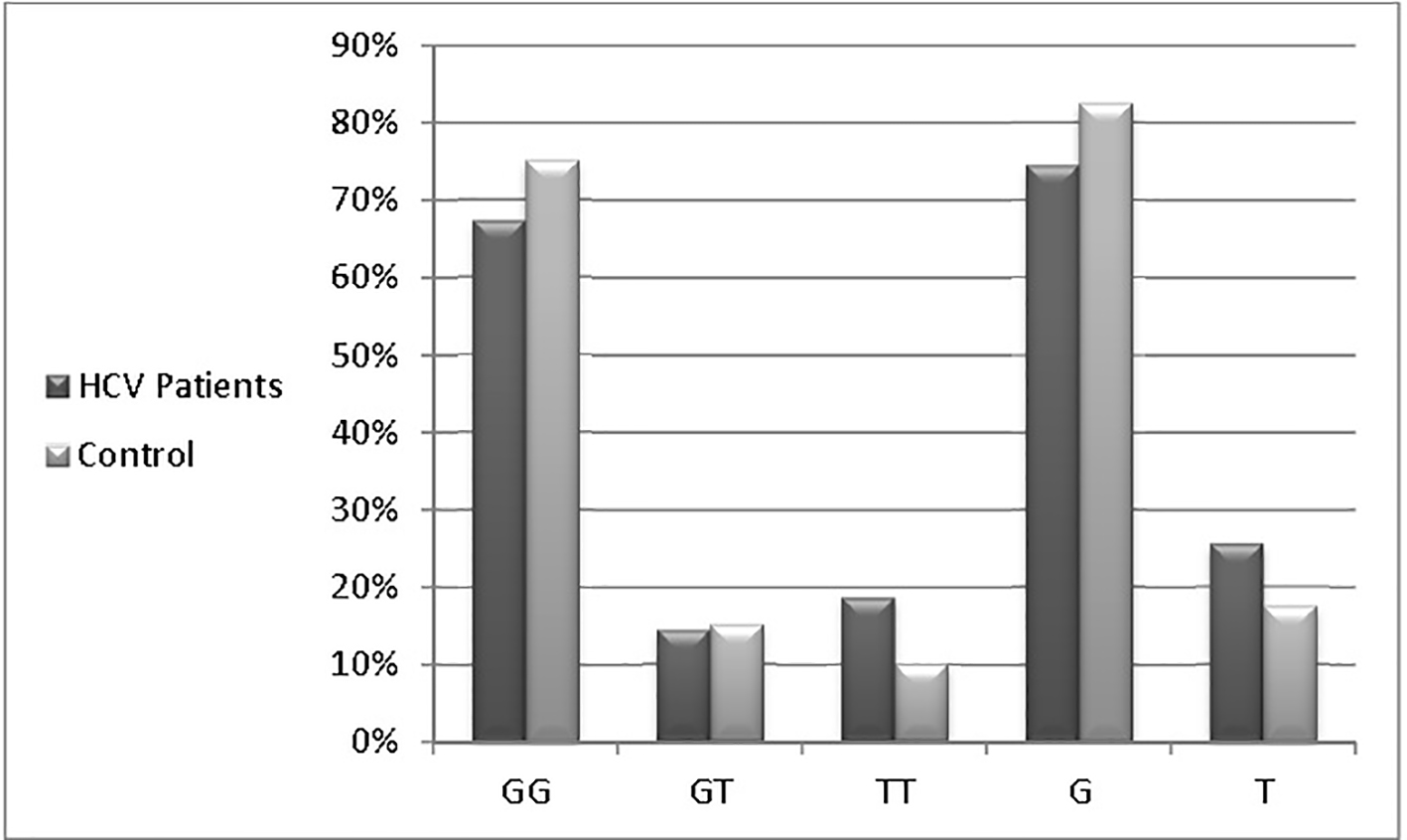 Click for large image | Figure 2. Genotyping and allele frequency of patients and control groups. |
 Click to view | Table 5. Treatment Responsiveness Parameters Between Both Studied Patients Groups |
 Click to view | Table 6. Correlation Between Predicting Factors and Treatment Responsiveness |
| Discussion | ▴Top |
Evolution of HCV treatment using DAAs is still not covering the whole countries, especially the resource limited regions rendering the treatment with PegIFN and RBV combination as one of the best solutions. Different ways were selected to augment the effectiveness of PegIFN treatment through adding one of DAAs members [10], using off-label indications of certain drugs [11] or even searching for the best predictors of treatment response [12]. Many circulating chemokines claimed to have an impact on fibrosis progression and response to PegIFN therapy [13, 14], also the major impact of IL28B gene polymorphism, which was used as one of the best predictors for either spontaneous viral clearance [15] or treatment outcomes [16]. MxA gene SNP at position -88 in the promoter region has a claimed influence on that gene expression and a possible relation to HCV response to treatment with PegIFN and RBV combination. In the current study, which included 70 chronic HCV patients undergoing antiviral therapy and 40 healthy controls, the univariate analyses showed no difference between those who showed treatment response or non-response regarding age (P = 0.092), ALT (P = 0.282), AFP levels (P = 0.669) and HCV viral load (P = 0.789). The carriers of different MxA genotypes did not show a remarkable difference either between HCV and control group (67% vs. 75%, 14.5% vs. 15% and 18.5% vs. 10% for GG, GT and TT genotypes, respectively) or HCV subgroups (non-responders and sustained responders) (76.7% vs. 60%, 13.3% vs. 15% and 10% vs. 25% for GG, GT and TT, respectively). In contrast, Bader El Din et al (2015) [17] reported a better sustained response to treatment in TT genotypes (66.7%) followed by GT (64.3%), and then GG (34.9%) in 60 chronic HCV genotype 4 patients. Similar results were reported by Shaker et al (2015) [18] in one study demonstrating that GG genotype was significantly lower in responders than in non-responders (25% vs. 75.76%), whereas heterozygotes GT were most likely responders (65.38% vs. 18.18%) in 85 chronic HCV genotype 4 patients. The pooled data from the previous studies showed the favorability of T allele carriers to achieve a better treatment outcome which is not clearly observed in our study in which the T allele was prevalent (25.5% vs. 17.5% in HCV and control groups, respectively). None of the studied populations had a well-established cirrhosis (F4); the worst response to treatment was seen in those with (F3) cirrhosis (0%). The results of our study call for larger studies or meta-analysis of all currently available data to confirm or refute the association of MxA SNP with response to PegIFN and RBV in patients with HCV genotype 4.
Conflicts of Interest
None of the authors have any financial conflicts of interest.
| References | ▴Top |
- Guerra J, Garenne M, Mohamed MK, Fontanet A. HCV burden of infection in Egypt: results from a nationwide survey. J Viral Hepat. 2012;19(8):560-567.
doi pubmed - Kandeel A, Genedy M, El-Refai S, Funk AL, Fontanet A, Talaat M. The prevalence of hepatitis C virus infection in Egypt 2015: implications for future policy on prevention and treatment. Liver Int. 2017;37(1):45-53.
doi pubmed - Muir AJ. The rapid evolution of treatment strategies for hepatitis C. Am J Gastroenterol. 2014;109(5):628-635; quiz 636.
doi pubmed - EASL Recommendations on Treatment of Hepatitis C 2016. J Hepatol. 2017;66(1):153-194.
doi pubmed - Gupta AC, Trehanpati N, Sukriti S, Hissar S, Midha V, Sood A, Sarin SK. Interleukin-28b CC genotype predicts early treatment response and CT/TT genotypes predicts non-response in patients infected with HCV genotype 3. J Med Virol. 2014;86(4):707-712.
doi pubmed - Landis H, Simon-Jodicke A, Kloti A, Di Paolo C, Schnorr JJ, Schneider-Schaulies S, Hefti HP, et al. Human MxA protein confers resistance to Semliki Forest virus and inhibits the amplification of a Semliki Forest virus-based replicon in the absence of viral structural proteins. J Virol. 1998;72(2):1516-1522.
pubmed - Fernandez M, Quiroga JA, Martin J, Herrero M, Pardo M, Horisberger MA, Carreno V. In vivo and in vitro induction of MxA protein in peripheral blood mononuclear cells from patients chronically infected with hepatitis C virus. J Infect Dis. 1999;180(2):262-267.
doi pubmed - Suzuki F, Arase Y, Suzuki Y, Tsubota A, Akuta N, Hosaka T, Someya T, et al. Single nucleotide polymorphism of the MxA gene promoter influences the response to interferon monotherapy in patients with hepatitis C viral infection. J Viral Hepat. 2004;11(3):271-276.
doi pubmed - Bedossa P, Poynard T. An algorithm for the grading of activity in chronic hepatitis C. The METAVIR Cooperative Study Group. Hepatology. 1996;24(2):289-293.
doi pubmed - Miuma S, Ichikawa T, Miyaaki H, Haraguchi M, Tamada Y, Shibata H, Taura N, et al. Efficacy and Tolerability of Pegylated Interferon and Ribavirin in Combination with Simeprevir to Treat Hepatitis C Virus Infections After Living Donor Liver Transplantation. J Interferon Cytokine Res. 2016;36(6):358-366.
doi pubmed - Esmat G, El Raziky M, Elsharkawy A, Sabry D, Hassany M, Ahmed A, Assem N, et al. Impact of vitamin D supplementation on sustained virological response in chronic hepatitis C genotype 4 patients treated by pegylated interferon/ribavirin. J Interferon Cytokine Res. 2015;35(1):49-54.
doi pubmed - El Raziky M, Fathalah WF, Zakaria Z, Eldeen HG, Abul-Fotouh A, Salama A, Awad A, et al. Predictors of Virological Response in 3,235 Chronic HCV Egyptian Patients Treated with Peginterferon Alpha-2a Compared with Peginterferon Alpha-2b Using Statistical Methods and Data Mining Techniques. J Interferon Cytokine Res. 2016;36(5):338-346.
doi pubmed - You CR, Park SH, Jeong SW, Woo HY, Bae SH, Choi JY, Sung YC, et al. Serum IP-10 Levels Correlate with the Severity of Liver Histopathology in Patients Infected with Genotype-1 HCV. Gut Liver. 2011;5(4):506-512.
doi pubmed - El Raziky M, Elsharkawy A, Said SE, Abdelatty S, El Akel W, Tantawy O, Gamal Eldeen H, et al. IP-10 Serum Level in Chronic Hepatitis C Virus Patients: Relation to Fibrosis and Response to Combined Interferon/Ribavirin Therapy. J Interferon Cytokine Res. 2015;35(8):649-653.
doi pubmed - Pedergnana V, Abdel-Hamid M, Guergnon J, Mohsen A, Le Fouler L, Theodorou I, Mohamed MK, et al. Analysis of IL28B variants in an Egyptian population defines the 20 kilobases minimal region involved in spontaneous clearance of hepatitis C virus. PLoS One. 2012;7(6):e38578.
doi pubmed - Matsuura K, Watanabe T, Tanaka Y. Role of IL28B for chronic hepatitis C treatment toward personalized medicine. J Gastroenterol Hepatol. 2014;29(2):241-249.
doi pubmed - Bader El Din NG, Salum GM, Anany MA, Ibrahim MK, Dawood RM, Zayed N, El Abd YS, et al. Association of Myxovirus Resistance Gene Promoter Polymorphism with Response to Combined Interferon Treatment and Progression of Liver Disease in Chronic HCV Egyptian Patients. J Interferon Cytokine Res. 2015;35(8):641-648.
doi pubmed - Shaker OG, Abdel-Rahim MT, Bayoumi ST. Gene polymorphisms of IL-10 and MxA in responders and non-responders to interferon therapy in HCV Egyptian patients genotype 4. Cell Biochem Biophys. 2015;71(2):617-625.
doi pubmed
This article is distributed under the terms of the Creative Commons Attribution Non-Commercial 4.0 International License, which permits unrestricted non-commercial use, distribution, and reproduction in any medium, provided the original work is properly cited.
Gastroenterology Research is published by Elmer Press Inc.


